6

















| Thumbs Up/Down |
| Received: 2,469/42 Given: 2,427/88 |


Now that IllustrativeDNA introduced QAPDM service, I thought starting a thread where everyone shares QAPDM models would be resourceful.
As a start, I am sharing this model:
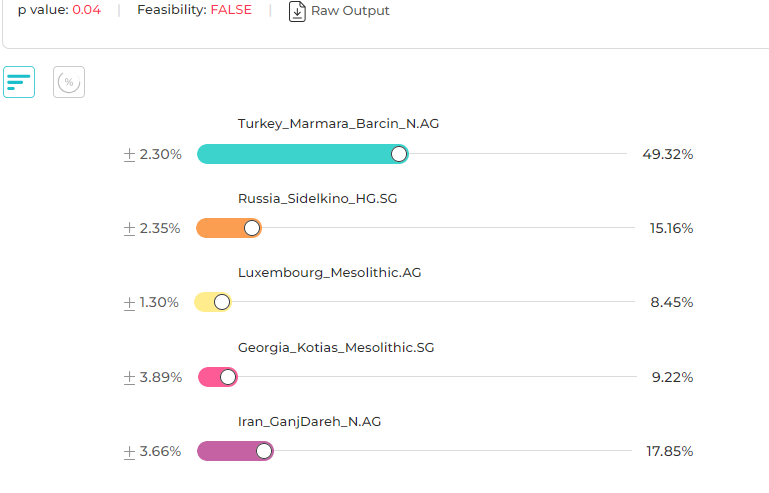
Sources
Turkey_Marmara_Barcin_N.AG (Anatolian Neolithic Farmer)
Russia_Sidelkino_HG.SG (Eastern European Hunter-Gatherer)
Luxembourg_Mesolithic.AG (Western European Hunter-Gatherer)
Georgia_Kotias_Mesolithic.SG (Caucasus Hunter-Gatherer)
Iran_GanjDareh_N.AG (Neolithic Iranian Farmer)
References
Mbuti.DG
Russia_AfontovaGora3_UP.AG
Italy_Epigravettian.SG
Russia_Kostenki14_UP.SG
Turkey_Central_Pinarbasi_Epipaleolithic.AG
Turkey_Central_Boncuklu_PPN.AG
Russia_Tyumen_HG.AG
Israel_Natufian.AG
Morocco_Iberomaurusian.AG
Georgia_Satsurblia_LateUP.SG
Iran_Wezmeh_N.SG
India_GreatAndaman_100BP.SG













| Thumbs Up/Down |
| Received: 818/19 Given: 341/54 |


Remove kostenki and Andaman from your rights. They're irrelevant to the model and will lead to worse pval
--














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |














| Thumbs Up/Down |
| Received: 779/0 Given: 221/1 |


I seem to get red p-value and feasibility
Y-DNA: I-Y3548 *updated*
mtDNA: H2a2a1














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


Testing the near east spice I see in G25 and Eurogenes. I have had AndreiDNA run this 2-way combination. I’m testing it myself with I believe a slightly different set of right populations. I also still cannot use my merged file, so I’m using my DNA file from MyHeritage.com. One is allowed two free kit uploads. It’s visible in the list, however, I cannot select it. I used the Human Origins Dataset for this.














| Thumbs Up/Down |
| Received: 17,471/170 Given: 13,083/176 |


I've been trying some medieval models but they keep getting rejected. Any tips how to improve?














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


Another model run with EGY_TIP_JK2134:
Sweden_IA_2 VK522:
Raw Output:
pat wt dof chisq p f4rank Sweden_IA_2.SG_VK522.SG Egypt_ThirdIntermediatePeriod.AG_JK2134.AG feasible best dofdiff chisqdiff p_nested
00 0 13 17.260705192920177 0.18765859345185965 1 0.8362559026573537 0.16374409734264636 TRUE NA NA NA NA
01 1 14 29.014137137307152 0.010404434189942862 0 1 NA TRUE TRUE 0 -43.42078257238162 1
10 1 14 72.43491970968877 6.968636070378395e-10 0 NA 1 TRUE TRUE NA NA NA
Denmark_IA VK532:
pat wt dof chisq p f4rank Denmark_IA.SG_VK532.SG Egypt_ThirdIntermediatePeriod.AG_JK2134.AG feasible best dofdiff chisqdiff p_nested
00 0 13 11.39641196621108 0.5776461449763756 1 0.8878444657143866 0.11215553428561346 TRUE NA NA NA NA
01 1 14 15.971954105271262 0.31509001573731876 0 1 NA TRUE TRUE 0 -56.16501842952157 1
10 1 14 72.13697253479283 7.896541490264353e-10 0 NA 1 TRUE TRUE NA NA NA
Scotland_MIA I2982:
pat wt dof chisq p f4rank Scotland_MIA.AG_I2982.AG Egypt_ThirdIntermediatePeriod.AG_JK2134.AG feasible best dofdiff chisqdiff p_nested
00 0 13 21.56134889984603 0.062542307243352 1 0.8882680436329078 0.11173195636709223 TRUE NA NA NA NA
01 1 14 25.82020873179249 0.027282545571266602 0 1 NA TRUE TRUE 0 -48.52234493063585 1
10 1 14 74.34255366242834 3.1231520993584695e-10 0 NA 1 TRUE TRUE NA NA NA
I had AndreiDNA model me with Old Kingdom Egyptian (NUE001) + Denmark IA and from there I copied the reference populations he used in that model run:
I had to substitute Russia_Karelia_HG to Russia_Samara_HG. For some reason I couldn't find it in IllustrativeDNA.
Also, I'm not sure If he did this, but I pooled together a number of reference populations into one. For example, I combined Taforalt 9-13 into the Iberomaurusian reference:
Full reference list:

There are currently 2 users browsing this thread. (1 members and 1 guests)
Bookmarks