3





















| Thumbs Up/Down |
| Received: 360/0 Given: 175/0 |


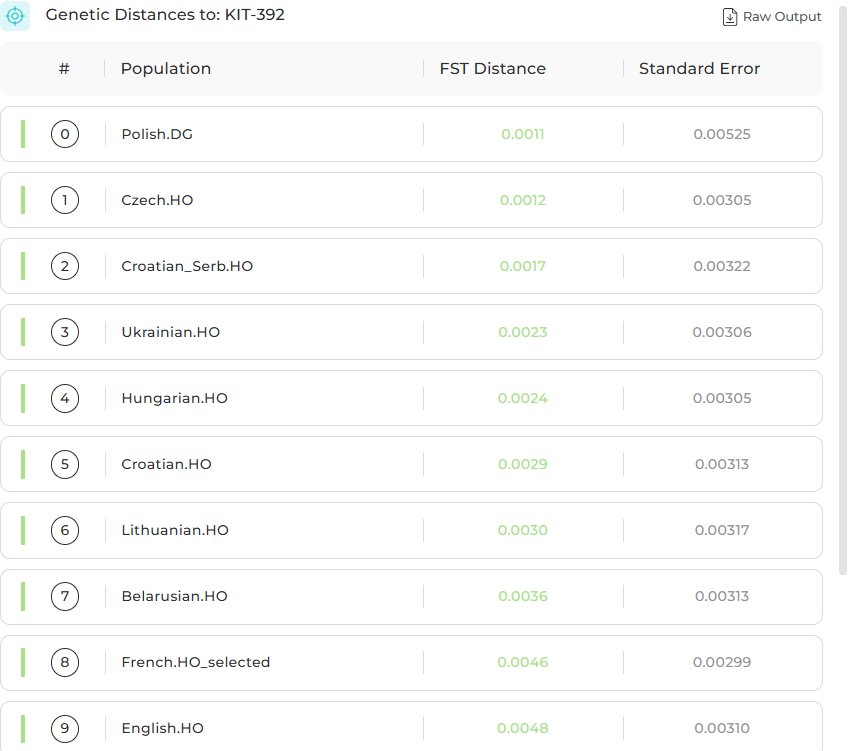
Some of my Genetic distances













| Thumbs Up/Down |
| Received: 818/19 Given: 341/54 |

















| Thumbs Up/Down |
| Received: 360/0 Given: 175/0 |
















| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |

















| Thumbs Up/Down |
| Received: 17,159/194 Given: 8,106/117 |


I'm a noob. Is there some method or reasoning to choosing the right reference populations?
Spoiler!













| Thumbs Up/Down |
| Received: 818/19 Given: 341/54 |


A. If you want to model using lefts that are real/exist, you select rights that are ancestral to your lefts, or if you can't find those that are ancestral, you have to search for those closely related to your lefts.
An example of that would be
your lefts:
EHG
WHG
ANF
CHG
then your rights have to be:
WHG
ANE (Ancestral to EHG)
ANF/you can break it down into AHG + Natufian + iran N
CHG
If you want to model
lefts:
ANE
WHG
Dzudzuana
You'd have to select rights that are ancestral such as
WHG
Yana (Ancestral to ANE), or alternatively something east eurasian like TY (Which is also ancestral to ANE)
Alternative Dzudzuana right, or
Natufian (Will work since dzudzuana can fit as a intermediate between Natufian, which is a right, and WHG, which is a right)
B. If you want to model with a ghost population, you select rights affinity to which could infer affinity to this ghost population, and the rest of rights you do as in point A.
An example of that would be modeling for basal eurasian admix. Since basal eurasian admix would decrease the affinity to neanderthals and denisovans, those have to be in the rights. Then you follow point A and chose lefts and rights for west eurasians and east eurasians.
--

















| Thumbs Up/Down |
| Received: 542/63 Given: 268/40 |


Hey, I just started out too — I’ve been using the qpAdm interface on Illustrative DNA, which makes it a lot more accessible for beginners.
As for choosing references ("right pops"), it’s a mix of PCA-based reasoning, historical context, and trial-and-error. I break it all down in a thread I posted here:
➡️ �� Stable qpAdm Models of Ashkenazi Jewish Ancestry – Ignoring PCA No More
Would love to chat more over there — I put a lot of effort into the models and framework and it barely got a reply, lol. Feel free to ask anything in that thread. Happy to help however I can.














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


My Dzudzuana + ANE admixture:
References:
I revisited a model AndreiDNA made for me where I was modeled as 3 way mix of Dzudzuana, Italy Epigravettian, and Afontovagora3 (ANE)
Chi-squared= 6.6
pat wt dof chisq p f4rank Georgia_Dzudzuana_UP.AG_I11857.AG Italy_Epigravettian.AG Russia_AfontovaGora3_UP.AG feasible best dofdiff chisqdiff p_nested
000 0 7 6.604471680174148 0.47118912498381044 2 0.6190138010869063 0.23446826784160432 0.1465179310714894 TRUE NA NA NA NA
001 1 8 11.519608968168374 0.17395876562151708 1 0.7146053472659971 0.2853946527340029 NA TRUE TRUE 0 -0.6103912843959698 1
010 1 8 12.130000252564344 0.1454971638521046 1 0.7886224100986546 NA 0.21137758990134542 TRUE TRUE 0 -87.06245602662968 1
100 1 8 99.19245627919402 6.242381618212807e-18 1 NA 0.21391679165755226 0.7860832083424478 TRUE TRUE NA NA NA
011 2 9 18.588268854606014 0.0289304686658288 0 1 NA NA TRUE NA NA NA NA
101 2 9 569.2036842919447 8.482679409878293e-117 0 NA 1 NA TRUE NA NA NA NA
110 2 9 103.07209455485025 3.756961650459967e-18 0 NA NA 1 TRUE NA NA NA NA
For the result on IllustrativeDNA (First Image), I used AndreiDNA's rights and added Ethiopia 4500 BP (Mota) to the references:















| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


Hey Andrei, I was modeling Morocco_MN skh001 and had some interesting results:
References:
Takarkori isn't available on IllustrativeDNA. I was wondering if you could try this model with Takarkori instead of Iberomaurusian. And try it with Iberomaurusian absent from the references and in. I want to see if skh001 is going to score around 10% Takarkori in this setup and the 16% or so assigned to Natufian.

















| Thumbs Up/Down |
| Received: 542/63 Given: 268/40 |


PCA vs. qpAdm: More Similar Than People Think
Fundamentally, autosomal PCA and qpAdm modeling operate on the same basic idea:
They both assess how populations are related based on genome-wide allele frequencies — just using different statistical approaches.
- PCA uses principal components to reduce genetic variance into 2D/3D plots, revealing clustering patterns.
- qpAdm estimates ancestry proportions by modeling a test population as a mix of chosen “left” sources, with “right” reference groups to anchor the model.
But in both cases, your results depend almost entirely on your inputs.
You can cherry-pick populations in either method — and get misleading results that still “look” scientific.
Why qpAdm Isn't Immune to Narrative Bias
There’s a tendency to treat qpAdm as more rigorous than PCA, and while it can be more formalized, it’s just as vulnerable to biased modeling choices.
That’s why so many published studies claim Ashkenazi Jews are “50%+ Levantine” — even though PCA consistently places them overlapping with Southern Italians.
Here’s why:
Most of these studies use deeply flawed European proxies — Northern Italians, French, Sardinians, or even Tuscans — while completely ignoring Southern Italians or Sicilians. This artificially inflates the Levantine component, because these proxies are genetically too distant and don't match PCA placement.
The qpAdm model still returns a “feasible” result — because qpAdm doesn't know your sources are bad.
It just fits whatever you give it.
In other words: Bad source selection in qpAdm doesn’t get corrected by the math — it gets rewarded. The tool will give you a p-value and ancestry breakdown that looks clean, but is fundamentally built on cherry-picked assumptions.
PCA-Informed qpAdm is the Solution
That’s why my models start with PCA — which clearly shows Ashkenazi Jews clustering directly on top of Southern Italians — and then use qpAdm to test models that reflect that structure.
Examples from my thread:
Stable qpAdm Models of Ashkenazi Jewish Ancestry – Ignoring PCA No More
Models such as:
- Italian_South.HO + Lebanese_Muslim.HO
- Italian_South.HO + Israel_Ashkelon_LBA.AG
- Italian_South.HO + Israel_C_o2.AG
All produce stable 2-way models with:
- ~75–78% Southern Italian–like ancestry
- ~22–25% Levantine
- Strong p-values and standard errors
- Full consistency with PCA and historical records
This matches not just the genetic structure, but also the well-documented Roman and Byzantine-era Jewish presence in Southern Italy.
Visual Results

Modern Levantine Proxy model

Bronze Age Coastal Levantine (Philistine-admixed)

Inland Bronze Age Levantine (likely Israelite/Canaanite)
Don’t Let Narrative Modeling Skew the Data
You can go check nearly any study by Behar, Hammer, or Ostrer — they almost all use Northern Italians or French as the European side of the model.
Even YouTubers like Andrei DNA — who is also an Apricity forum member — repeat the same setup and get the same distorted results: Ashkenazi Jews as a “Middle Eastern” population.
But if Ashkenazi Jews sit right on top of Southern Italians in PCA — and they do — then they’re no more “Middle Eastern” than a Calabrian is.
That’s why PCA-aligned qpAdm is essential — and narrative-driven modeling needs to be called out.
Last edited by Lard; 07-20-2025 at 03:51 AM.
There are currently 2 users browsing this thread. (0 members and 2 guests)
Bookmarks