0

















| Thumbs Up/Down |
| Received: 13/1 Given: 0/0 |


I know Dienekes didnt recommented using Oracle on tiny DNA fragments instead of genomewide but I wanted to know what would be the result anyways.
I flushed every single Chromosom through the Oracle thing and these are the top nation for each Chromosom:
(only Nations and no doublettes)
Chromosom 1: Orkney Island
Chromosom 2: Slovenia
Chromosom 3: Germany
Chromosom 4: Sweden
Chromosom 5: Germany
Chromosom 6: Slovenia
Chromosom 7: Germany
Chromosom 8: Finland
Chromosom 9: Germany
Chromosom 10: Netherland
Chromosom 11: Finland
Chromosom 12: France
Chromosom 13: Germany
Chromosom 14:Finland
Chromosom 15: Slovenia
Chromosom 16: Finland
Chromsom 17: France
Chromosom 18: Germany
Chromosom 19: Germany
Chromosom 20: Germany
Chromosom 21: Norway
Chromosom 22: Italy
8/22 Germany
4/22 Finland (because of my Lithuanian Great Great Grandmom maybe)
3/22 Slovenia
2/22 France
1/22 Orkney Island
1/22 Norway
1/22 Sweden
1/22 Netherland
1/22 Italy
EDIT:
Needed to Edit, my Great Grandmom into my Great Great GRandmom... lol
Last edited by Frederick; 08-22-2011 at 10:17 PM.
Bring back the stocks!













| Thumbs Up/Down |
| Received: 84/1 Given: 27/1 |


I'd imagine that it is due to the different amounts of SNPs in your chromosomes. Chromosome 1 has 13376 SNPs, while chromosome 22 has but 2688. Consequently, Chromosome 1 influences the actual average to a much greater degree. Simply averaging by chromosome results will not take this into account, and hence will yield little.
Single Population Sharing (Eurogenes)
1 Estonian_Polish 4.36
2 Russian_Smolensk 4.88
3 Southwest_Russian 5.32
4 Belorussian 5.78
5 Lithuanian 5.92














| Thumbs Up/Down |
| Received: 2/0 Given: 0/0 |


Why does everyone get wog and chink scores on this, but on Eurogenes and previous Dienekes tests they get no non-Euro?













| Thumbs Up/Down |
| Received: 137/0 Given: 28/0 |


My "bychr."
Code:Chr E_Euro W_Euro Medite Neo_Af W_Asia S_Asia N_E_As S_E_As E_Afri S_W_As N_W_Af Pal_Af 1 2.57 0.37 23.6 0 52.23 0 1.97 0 0.01 19.25 0 0 2 2.43 7.58 22.79 0 43.63 0.02 0 0 0 23.54 0.02 0 3 1.34 2.82 31.18 0.02 45.32 9.02 0 0 0.36 8.92 1.02 0 4 0.01 2.73 17.59 0.09 50.22 11.48 1.04 0 0 16.84 0 0 5 0 4.67 31.83 0 30.77 5.28 0.26 0 1.03 26.16 0 0 6 6.09 0.04 23.46 0 44.85 0.56 2.06 0 0 21 1.94 0 7 0.07 0 28.7 0.32 45.66 3.18 0.28 0.74 2.85 18.21 0 0 8 0.03 0 27.38 0.3 49.83 2.77 0 0.73 0 15 3.96 0 9 4.26 6.9 23.51 0 38.02 10.47 0 0 0.75 16.09 0 0 10 8.41 1.35 16.76 0 41.03 9.85 0 1.95 0 17.47 2.28 0.89 11 0 0 21.4 0 50.13 0.01 0 0 0 21.33 7.12 0 12 4.96 6.81 20.94 0 44.62 0.37 0 0.17 0 22.13 0 0 13 0 0.04 23.8 0 46.45 10.43 0 5.86 0 12.84 0 0.58 14 1.47 2.49 14.57 0.01 44.33 12.26 1.66 0 0.01 20.57 1.45 1.16 15 0.01 11.11 23.39 0 51.67 0 0 0 1.05 10.51 2.25 0 16 0.02 4.23 31.48 0 41.27 5.44 0.17 0 0 17.38 0 0 17 0 18.13 25.62 0 36.75 2.32 0 0 0 17.15 0.02 0 18 0.03 0 20.58 0 39.15 0 0 3.58 3.03 33.63 0 0 19 0 0 24.13 0 42.68 4.25 0 2.13 5.94 19.18 1.69 0 20 5.33 0 29.48 0 41.21 5.54 0 3.95 0.3 14.18 0.01 0 21 7.88 0.01 19.45 1.58 44.59 4.45 4.19 2.48 0 15.36 0 0 22 0 14.23 24.56 0 34.45 2.79 0 0.13 0 23.84 0 0














| Thumbs Up/Down |
| Received: 36/3 Given: 0/0 |


I just received admixture analysis results from Dienekes. I am DOD804 on the spreadsheet. I sorted them.
Mediterranean 34.8
West_Asian 27.4
West_European 17.2
Southwest_Asian 11.4
Northwest_African 6.6
East_European 1.5
East_African 1.1
Neo_African 0.2
South_Asian 0
Northeast_Asian 0
Southeast_Asian 0
Palaeo_African 0














| Thumbs Up/Down |
| Received: 45/0 Given: 4/0 |


My DIY Dodecad2.0 results
Genome-wide results:
41.89% Mediterranean
41.69% West_European
5.50% Northwest_African
3.34% Southwest_Asian
3.27% West_Asian
1.81% East_African
1.24% East_European
0.63% Southeast_Asian
0.61% Neo_African
0.01% South_Asian
0.00% Northeast_Asian
0.00% Palaeo_African
By chromossome:
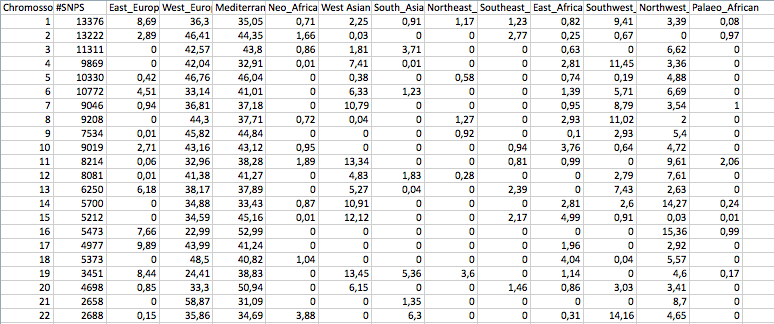













| Thumbs Up/Down |
| Received: 84/1 Given: 27/1 |


'FIN' and 'German_D' are Dodecad's primary way of explaining West Europeans with a significant East European element. Germans_D and FIN (not Finnish_D) are principally rather similar to each other, except that Finns are less Mediterranean.
25% EE, 55% WE and 20% Med = German_D, then FIN
25% EE, 55% WE, 10% Med, 10% scattered = FIN, then German_D
A hypothetical Prussian German would turn out FIN, for instance: he will be mostly WE, with a moderate EE and little Med. Lithuanians do have the single highest EE percentage of all nations there, but it's unlikely that such a far-away ancestor would be a significant influence.
Single Population Sharing (Eurogenes)
1 Estonian_Polish 4.36
2 Russian_Smolensk 4.88
3 Southwest_Russian 5.32
4 Belorussian 5.78
5 Lithuanian 5.92














| Thumbs Up/Down |
| Received: 1,828/89 Given: 1,086/83 |


My" bychr"
Everything else is to small to tell if it's noise or not.
There are currently 1 users browsing this thread. (0 members and 1 guests)
Bookmarks