0

















| Thumbs Up/Down |
| Received: 111/0 Given: 11/0 |


Closest ancient populations:
Viking + Svear (7.588)
Viking + Curonian (8.638)
Svear + Scythian (9.269)
Svear + Curonian (9.693)
Viking + Ostrogoth (9.9)
Svear (10.05)
Scythian (12.6)
Viking (13.21)
Curonian (14.38)
Ostrogoth (15.99)
Closest modern populations:
1. Finnish (3.020)
2. Southwest_Finnish (5.737)
3. East_Finnish (7.475)
4. Estonian (10.56)
5. North_Swedish (11.13)
6. German_Central (13.38)
7. Polish (13.56)
8. South_Polish (14.28)



| Thumbs Up/Down |
| Received: 13/0 Given: 0/0 |


Well the above were my DNAGenics coordinates. This is Illustrative DNA:
Christine__scaled,0.125205,0.131003,0.053551,0.027 455,0.0397,0.011713,0.00893,0.008307,0.002045,0.00 4009,-0.004709,0.005095,-0.005946,0.001927,0.011808,-0.004773,-0.01682,0.004687,0.006913,-0.008629,0.001622,0.002473,-0.004067,0.009881,-0.000239



| Thumbs Up/Down |
| Received: 13/0 Given: 0/0 |



| Thumbs Up/Down |
| Received: 13/0 Given: 0/0 |



| Thumbs Up/Down |
| Received: 13/0 Given: 0/0 |


I have no such knowledge, no. My Heritage says 2.6% Iberian with an additional genetic group of "Mexicans in New Mexico and Colorado" with confidence level high. The common ethnicities for that genetic group is mostly Mesoamerican and Andean. I figure My Heritage must be mistaking a bit of Amerindian with some Iberian that I'm not aware of and combining it into Mexican American. Best guess.



| Thumbs Up/Down |
| Received: 13/0 Given: 0/0 |


So a lady that's helped me with the minority ancestry issue suggested I use some specific calculators at DNAGenics and a method of seeing what ancestry my results seem to gravitate to. So she said to check one each of what I know I am, such as German and English, then choose one northeast Asian, I choose Han Chinese, then check a bunch of Amerindian and see what it reveals. She said my result indicated likely Amerindian ancestry which she felt was confirmed by several ancient matches in the Genomelink Ancient Bloodlines report in the Americas. It's pretty confusing but interesting.
This is the type of result I'd get from the above settings at DNAGenics:
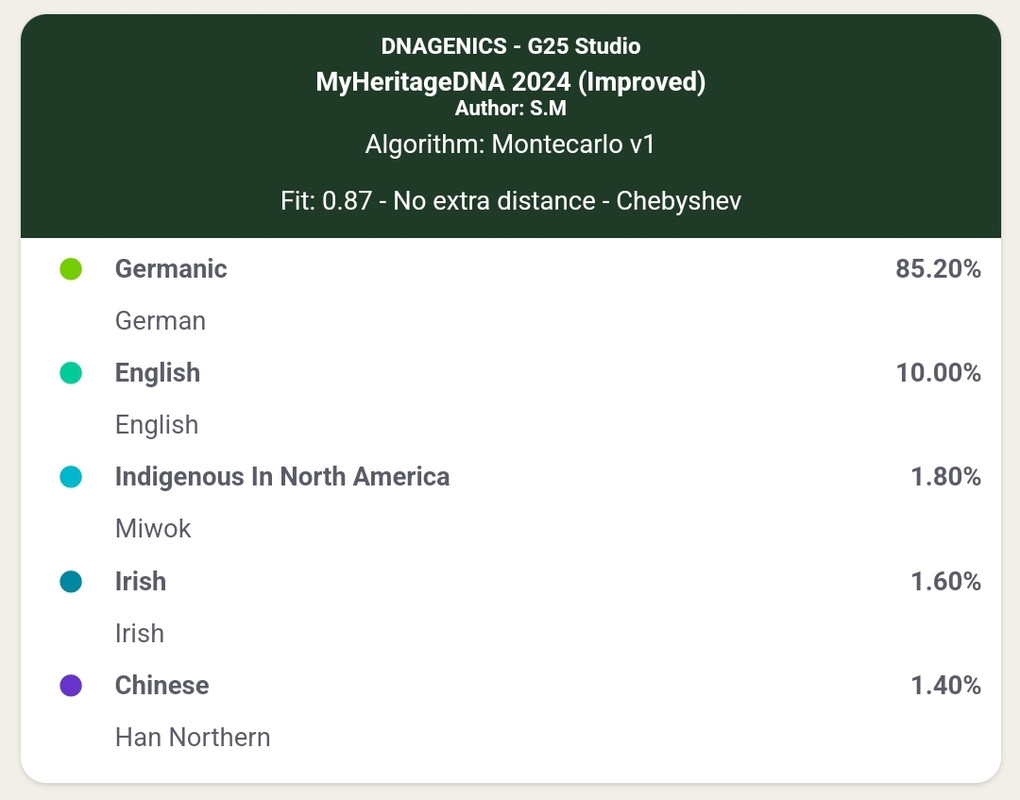














| Thumbs Up/Down |
| Received: 2,386/2 Given: 3,577/14 |


Target: Christine__scaled
Distance: 0.0215% / 0.02147271 | ADC: 0.5x RC
46.9 Germany
46.3 Belgium
6.8 Turkey
Target: Christine__scaled
Distance: 0.0193% / 0.01927877 | R4P
60.7 England
19.9 Italy
17.7 Belarus
1.7 Kirghizstan
Target: Christine__scaled
Distance: 0.0203% / 0.02030693 | R3P
66.3 England
18.3 Belarus
15.4 Israel
Hmm interesting. I'll add the Native American may just be a misread showing overlap with Ancestral North Eurasians which are a component to western steppe herders like the Yamnaya culture.



| Thumbs Up/Down |
| Received: 13/0 Given: 0/0 |


Well that's what I've tried to figure out. There is clearly some genetic overlap between Amerind and Eurasian. The point of putting Han Chinese into the DNAGenics calculator was to discern which side of the Pacific were the genetics pointing to.
Aside from that, my Genomelink ancient samples that point toward Amerind are:
Ancient Beringian Toddler 0.24%
Paleoindian Woman of the Andes 0.07%
Belizean Hunter-Gatherer 0.12%
Ancient Argentine Well Digger 0.06%
Archaic Peruvian Male 0.05%
Brazilian Potter Woman 0.06%
Medieval Age Caribbean Man 0.12%
I'd love to know the result of anyone from Europe or certified non-admixed European heritage of this test to see if they're getting anything like this.














| Thumbs Up/Down |
| Received: 2,386/2 Given: 3,577/14 |


I get native american matches on genomelink too:
California Native American Woman
SN-04
0.53% DNA match
Paleoindian Woman of the Maya Mountains
I3443
Ancient Beringian Toddler
Cuban Fisherman
GUY002
Which causes me to question if they have the algorithm "too loose" so to speak.



| Thumbs Up/Down |
| Received: 13/0 Given: 0/0 |
There are currently 1 users browsing this thread. (0 members and 1 guests)
Bookmarks