0


















| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


You have a bias as well. And it's against G25 which I cant really remember why. Except maybe your results on there don't fit your worldview on how people and places should be.
On your point with Satsurblia vs. Kotias. Kotias is closer in time to when CHG groups were interacting with EHGs in the steppe. And it makes more biological sense to use Kotias for European admixture modeling. Satsurblia is used for more deep time modeling. It's almost like using Dzudzuana in a neolithic/mesolithic model.
I essentially googled this because I think you are oversimplifying G25 and If I spend a whole hour or whatever writing my own defense of G25, it would be a waste of my time:
PCA basis isn’t inherently invalid – G25 coordinates are derived from a PCA built with ancient genomes, which is conceptually similar to the PCA space scientists use in publications. qpAdm, formal stats, and supervised modeling often rely on the same PCA framework for visualizing population structure.
Correlation with formal models While G25 is not a formal f-statistics tool, supervised models on G25 data can often produce admixture proportions that track closely with qpAdm, especially when the reference set is chosen well.
p-values aren’t the whole story Even in qpAdm, p-values in the 0.05–0.2 range can be considered reasonable, especially when reference populations aren’t perfect fits. Chasing a p=1.0 is unrealistic because even published academic models rarely achieve that without overfitting.
Scientists do use PCA-based coordinates – The difference is that in academic work they generate their own PCA from raw data, but the underlying principle is the same. The main limitation with G25 is that it’s a fixed PCA space and coordinates are produced by projection rather than recomputing the PCA, which can introduce some distortion—but it’s still valid for exploratory work.
I'd like to know what papers you are referring to. The ones I have seen when it relates to European genetics, it's Kotias. And Satsurblia is used for deeper time modeling of Kotias itself.
Last edited by Gannicus; 08-14-2025 at 05:38 PM.














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


Just like in G25 I can model myself with near eastern groups. As I mentioned before it's not a perfect exact match. Also, another difference I'm qpAdm modeling with MyHeritage DNA file and not my Merged DNA file that has my data from 23&Me v4, v5, and Ancestry.com.
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0217% / 0.02173807
96.1 English
3.9 Yemenite_Jew
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0230% / 0.02297868
93.9 Scottish
6.1 Lebanese_Druze
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0225% / 0.02251603
95.2 Scottish
4.8 Saudi
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0223% / 0.02226118
95.9 English
4.1 Samaritan
(only 1 Samaritan in AADR dataset vs this G25 average)














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


And here are some more:
Attachment 142650
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0225% / 0.02247160
95.2 Scottish
4.8 BedouinB
Attachment 142651
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0226% / 0.02261509
93.7 Scottish
6.3 Lebanese_Christian
Attachment 142652
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0239% / 0.02386469
95.0 Scottish
5.0 Assyrian
Attachment 142653
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0220% / 0.02195749
95.1 English
4.9 Lebanese_Christian














| Thumbs Up/Down |
| Received: 91/0 Given: 19/0 |


So that is all you can say i have a biase. It seems i hit a nerve. You go at my worldview ..... look i aprouch every nation as it's own and try to reasonably get to an answer after looking at every side. Just a simple explanation why i argue they way i do.
Now something i have noticed on this forum is that armchair geneticist only follow some low level claculations with g25 which is from the bodom completely biased. Davdski has an agenda that is a simple a fact!
I read alot and get my sources from papers and professionals themselves. And i care about unbiased truth. Wich Davidski doesn't provide since he hides his methodology which is unprofesional and adds to his amateurish ways. So you seem to defend him for the life of it which is really weird.
You seem to know more than actual geneticist ? Anyway those Qpadm results confirm everything i got from my DNA report.
I am going to repeat yet again G25 is only good for general clustering nothing more.
Now let's come to your argument with Kotias and Satsurblia. This "Kotias fits the timeline better" argument is an oversimplification, and in population genetics, time alone is not the main reason to pick a proxy. Snps don't carry a timeline or exact date!
The goal in QPADM is to represent the ancestral component, not to match the exact migration date!
If an older sample like Satsurblia is genetically closer to the pure CHG source then it is the better choice simple as that. Which P value and standard error prooves.
Many major papers like Lazaridids , Mathieson and Narasimhan etc just to name a few use Satsublia for CHG whne they want to avoid steppe bias , even modeling past 9000 BP populations.
Point is it is about what fits your genetic composition th e best that is why each snp has to be looked at. Where G25 tries to pressure your raw data into 25 dimensional points which creates heavy noise and very unprecise results when it comes to ancestral composition that is a fact and every academic will tell you the same!
So if you come only with a google search answer this tells me already your approach to things, but whatever.
Think about that , this is no attack on you or anything , don't wanna start an unecessaryy fight.
If you wanna know ,yes G25 get´s it correctly in clustering me to Croats/Slovenians/Bosniak and Hungarians. Everything else ..... well i explained myself enough.
Kind regards














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


I hit a nerve of yours earlier when I was using citing G25 and Eurogenes k13. The only reason I defend it is because I see similarity with qpAdm and other tools. I clearly demonstrated that there is correlation with G25 and qpAdm in my previous posts, however, I also acknowledge that it doesn't always line up. So your position on G25 doesn't necessarily hold up. From your response I could tell right away you didn't like your results and that It does not align with your perceived ancestral background. Or at least that's the vibe I got from the way you worded your post.
It certainly sounded like that based on the responses I get out of you. It sounds exactly like someone who likes to put people in specific boxes and if the data doesn't fit then its wrong. I didn't outright reject your model with Satsurblia either. Just a suggestion that you should try Kotias. I'll say that maybe I misinterpreted some of your posts. And if you do substitute Satsurblia for Kotias and pval drops to 0.1 or something, that is still valid.So that is all you can say i have a biase. It seems i hit a nerve. You go at my worldview ..... look i aprouch every nation as it's own and try to reasonably get to an answer after looking at every side. Just a simple explanation why i argue they way i do.
You seem to know more than actual geneticist ?
Did I say I do? No, neither do you. Your tone could use some work. You came in here being dismissive and mildly annoyed in the first place. Your opinion on Davidski and how you worded it draws suspicion to you. And I can't quite place my finger on it but it's an attitude I have seen on here where members sh*t on everyone on here despite the fact he/she is a member and participating here.
Also:
So if you come only with a google search answer this tells me already your approach to things, but whatever.^You seem to know more than actual geneticist ?lol?this is no attack on you or anything , don't wanna start an unecessaryy fight.
Edit:
I can't really take anything you say seriously after this comment:
Excuse my French, but that is total bullshit.I might be to harsh but i would only take p values around 0,5 and above serious.
Last edited by Gannicus; 08-14-2025 at 09:45 PM.














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


Another Steppe+EEF/ANF+WHG model. I have modeled myself with Germany LBK already, but that was with a Corded Ware group.
Target: Gannicus_MergedFile_officialDavidski_scaled
Distance: 0.0439% / 0.04392507
46.8 Russia_Samara_EBA_Yamnaya
43.1 Germany_EN_LBK
10.1 Luxembourg_Loschbour.DG
The admixture of Germany LBK EN:
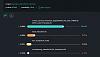
Last edited by Gannicus; 08-15-2025 at 04:55 AM.














| Thumbs Up/Down |
| Received: 91/0 Given: 19/0 |


Stop imagining what you can read out of my post ;your assumbtions are redicoulus. You even started to go personal trying to talk about my worldview which you have no clue of. Go to my beginning post i always kept it on topic always talking about the results and pointing towards professional sources.
But here you act as if you know better then them which proofs my assumptions right. And then you keep larping about me being dissadisifed with davidksi which is justified. Not because my G25 coords are somehow bad whcih is again wrong or as you frame it "it doesn't fit my worldview" which again ingonrant.
Proofs again that i hit a nerve just look how you started to respond. I didn't do the same.
If you cannot handle the truth then it is not on me.
Yes i can also reproduce all the Davidski G25 results but they fail or are extremly bad with high standard errors. Which gives me another inside on what he did or his methodology. Each and every geneticist approches population genetics as individuals or if there is a common ground then you go universal but with coution.
Everything i explained to you with CHG Kotias and satsurblia is perfectly valid (and you can look em up) just because you are ingorant and doesn't want to except that fact points towards your narrow view on things.
And yes the goal is to go well aboive a p-value 0,05 and keep the standard error low which in my case i provided.
Keep your manners in, don't try to assume anything about me i haven't attacked you previously.
And i am gonna repeat yet again G25 is ony good for general clustering. NOT FOR DEEP ANALYSIS
So let's go back to the topic this thread is about QPADM and them models . If you wanna argue with me write me personally.
Cheers!














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


I don’t think I act as if I know better than them. You’re just saying that because I don’t agree with you. Or it at least that’s what it seems like. I study this stuff in my free time. I’m always learning. And so are you. I only escalated because your tone changed. And I don’t fully agree with your positions on things. I was neutral/polite with you this entire time until I was saying things that you did not agree with. I went back and looked by the way. I’m very sensitive to tone. And if you have disagreement with someone about something like G25 you do not word your posts the way you did. It sounds like you have it all figured out too. Using language like “nonsense” from a random person on the internet that isn’t an authority on genetics is off putting. That was only one example. I reciprocate what I receive. And if you respond the way you did you will get what you got from me. My position is that G25 isn’t as bad as you say it is. It’s not perfect, but that doesn’t mean it should be thrown in a metaphorical trash can either.














| Thumbs Up/Down |
| Received: 91/0 Given: 19/0 |


Look you are using one word examples to as an escuse to attack me personally. You know that you have crossed the line. I kept it on topic you went for personal stuff which you have no clue of.
Yes you do act as if you know just by judging how you wrote and attacked me. And you should stop imagining things how someone sounds over a text you seem a little emotional. So i will ask you kindly yet again to keep your manners in.
I have provided enough info on the qpadm vs g25 thing and the whole genetics analysis, where you did not. You proved lazy google search answers.
Yes i do read alot about that topic and if you would to and be more critical and less emotional and just go for the facts you will see what i point to.
And if you look close enough i pointed out way early that G25 is only good for general clustering/distances to poluations nothing more nothing less.
So i have a clear view and opinion on that but your emotions and some fanatsys clouded that the whole thing.
Anyway the problem is sovled let's move back to the general topic which is QPADM!














| Thumbs Up/Down |
| Received: 2,454/2 Given: 3,699/14 |


I just posted one example. You want me to link the post and highlight your shitty word choice? I’ve been biting my tongue since you made that ridiculous comment about p-values. Claiming I don’t review or read the papers when I actually linked a paper that refuted your bullshit statement. Your tone was shit and you are the one calling everyone on here “amateurs”. Who the fu*k are you to be saying shit like that?? I have built up a pretty good knowledge base. And there is always room to learn more. But the way you spoke in a couple posts I cannot let that stand. I have absolutely 0 patience for condescension. I’ve let it slide in the past but not anymore. That's why you are getting this. I'm done with it. Even if it's a little bit. You know why? Because it's incredibly f*cking easy to be neutral/polite.
Also, just to make you happy do I have to put a disclaimer out every time I make a post that says I'm not a population geneticist, there's gaps in my knowledge, and I'm still learning?? Because nobody does that. And another thing, I have interacted with other members on here. Majevica corrected me in this thread and suggested I use a different EHG in my references for qpAdm. Do I think he is a know it all because of that? Absolutely not! It helped me out.
Your tone was garbage in the first place and now you are acting like you're the righteous one after I had enough? That's rich.
Last edited by Gannicus; 08-16-2025 at 01:40 AM.
There are currently 1 users browsing this thread. (0 members and 1 guests)
Bookmarks